Supported hybrid-capture enrichment panels
Supported amplicon primer schemes on Protocols.io or ARTIC Network
- Universal Influenza A/B (Zhou et al / Ying Lin)
- MPXV (Grubaugh Lab)
- Dengue 400 bp (Grubaugh Lab)
- RSV (Barr Lab / Ying Lin)
- SARS-CoV-2 ARTIC v3, v4, v4.1, v5.3.2(ARTIC Network)
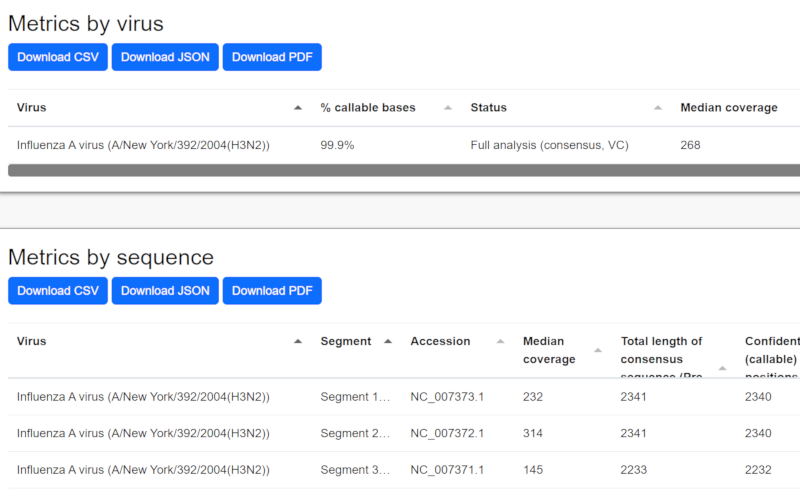
Analysis Pipeline
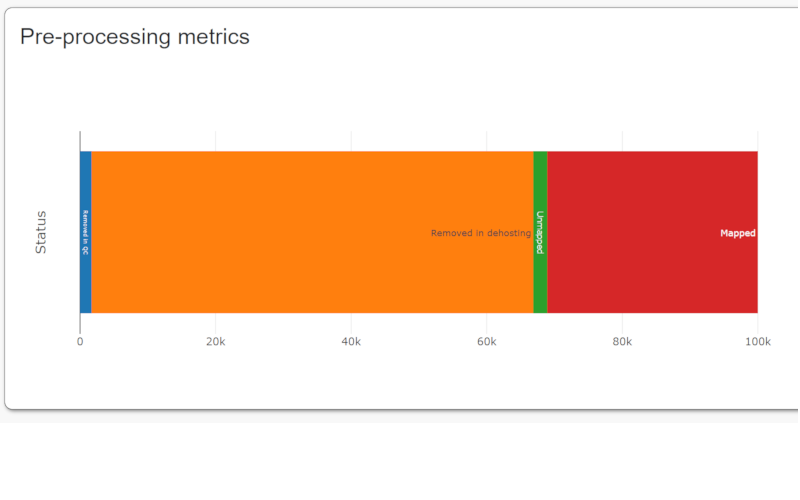
- Reads are trimmed and filtered using Trimmomatic with the following parameters: LEADING:3 TRAILING:3 SLIDINGWINDOW: 4:15 MINLEN:36.
- Human reads are removed with a modified version of the SRA Human Read Scrubber tool.
- MEGAHIT is used to perform de novo assembly on the scrubbed reads.
- CD-HIT-EST is used to cluster similar contigs to reduce redundancy.
- The resulting contigs are mapped to a set of reference genomes using minimap2.
- The best matching reference for each contig is selected for short read mapping.
- The scrubbed reads from step 3 are aligned to the selected reference genomes using [DRAGEN v4.0.3]
- Sequence variants are called from the alignments using DRAGEN Somatic v4.0.3 and applied to the corresponding reference sequences to create consensus sequences.
- If applicable, Pangolin and/or Nextclade are run on the consensus sequences.
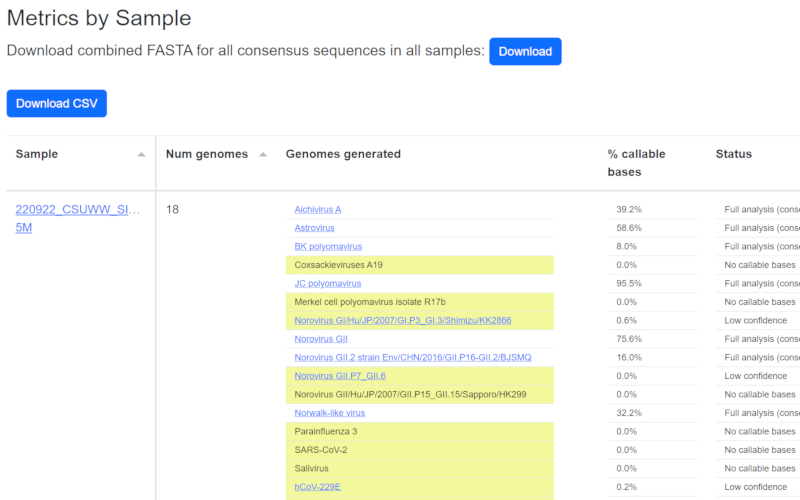
Outputs
The software generates sequences representing a best estimate of the population of targeted sequences in the sample. NextClade and Pangolin analysis are run on select organisms. Output details in output txt file.